Model Stacking¶
Here you’ll find the explanation about how to fit stacking models using aikit.
You can also see the notbook :
Stacking How To¶
Stacking (or Blending) is done when you want to aggregate the result of several models into an aggregated prediction. More precisely you want to use the predictions of the models as input to another blending model. (In what follows I’ll call models the models that we want to use as features and blender the model that uses those as features to make a final predictions)
To prevent overfitting you typically want to use out-sample predictions to fit the blender (otherwise the blender will just learn to trust the model that overfit the most…).
To generate out-sample predictions you can do a cross-validation:
for fold (i) and a given model :
- fit model on all the data from the other folds (1,…,i-1,i+1, .. N)
- call fitted model on data from fold (i)
- store the predictions
If that is done on all the folds, you can get predictions for every sample in your database, but each prediction were generated using out sample training set.
Now you can take all those predictions and fit a blender model.
StackerClassifier¶
This class does exactly what is describeb above. It takes as input a list of un-fitted models as well as a blending model and will generate a stacking model. In that case what is given to the blending model are the probabilities of each class for each model.
- class
aikit.models.stacking.StackerClassifier(models, cv, blender, random_state=None)¶generic class to handle stacking
This class takes a list of models and does the following during its fitting phase
- does a cross-validation on each model to output out-sample predictions
- use those out-sample prediction to fit a blending model
- re-fit the models on all the datas
During test: 1. call each models to retrieve predictions 2. call the blender to retrieve final aggregated prediction
Parameters:
- models (list of model) – the models that we want to stacked
- cv (cv object or int) – the cross-validation to use to fit the blender
- blender (model) – the blending model
StackerRegressor¶
Same class but for regression problems
- class
aikit.models.stacking.StackerRegressor(models, cv, blender, random_state=None)¶generic class to handle stacking
This class takes a list of models and does the following during its fitting phase
- does a cross-validation on each model to output out-sample predictions
- use those out-sample prediction to fit a blending model
- re-fit the models on all the datas
During test: 1. call each models to retrieve predictions 2. call the blender to retrieve final aggregated prediction
Parameters:
- models (list of model) – the models that we want to stacked
- cv (cv object or int) – the cross-validation to use to fit the blender
- blender (model) – the blending model
OutSamplerTransformer¶
The library offers another way to create stacking model. A stacking model can be viewed as another kind of pipeline : instead of a transformation it just uses other models as a special kind of transformers. And so GraphPipeline can be used to chain this transformation with a blending model.
- To be able to do that two things are needed:
- a model is not a transformer, and so we need to transform a model into a transformer (basically saying that the :function:`transform` method should use :function:`predict` or :function:`predict_proba`)
- to do correct stacking we need to generate out-sample predictions
The OutSamplerTransformer does just that. It takes care of generating out-sample predictions, and the blending can be done in another node of the GraphPipeline (see example bellow)
- class
aikit.models.stacking.OutSamplerTransformer(model, cv=10, random_state=123, desired_output_type=None, columns_prefix=None)¶This class is used to transform a model in a transformers that makes out of sample predictions
This transformation can be used to easily use model in part of a GraphPipeline.
fit method : 1. simply fit the underlying model
fit_transform method : 1. do a cross-validation on the underlying model to output out-of-sample prediction 2. re-fit underlying model on all the data
transform method : 1. just output prediction of underlying model
Parameters:
- model (a model) – the model that we want to ‘transform’
- cv (cv object or int) – which crossvalidation to use
- random_state (int or None) – specify the random state (to force the CV the be fixed)
- desired_output_type (None or type of output) – the output type of the result of the transformation
- columns_prefix (None or str) – each column will be prefixed by it
Example¶
Let’s say we want to stack a RandomForestClassifier, an LGBMClassifier and a LogisticRegression. You can create the model like that:
stacker = StackerClassifier( models = [RandomForestClassifier() , LGBMClassifier(), LogisticRegression()],
cv = 10,
blender = LogisticRegression()
)
and then fit it as you would a regular model:
stacker.fit(X, y)
Using OutSamplerTransformer we can achieve the same thing but with a Pipeline instead:
from sklearn.model_selection import StratifiedKFold
from aikit.models import OutSamplerTransformer
from aikit.pipeline import GraphPipeline
cv = StratifiedKFold(10, shuffle=True, random_state=123)
stacker = GraphPipeline(models = {
"rf" : OutSamplerTransformer(RandomForestClassifier() , cv = cv),
"lgbm" : OutSamplerTransformer(LGBMClassifier() , cv = cv),
"logit": OutSamplerTransformer(LogisticRegression() , cv = cv),
"blender":LogisticRegression()
}, edges = [("rf","blender"),("lgbm","blender"),("logit","blender")])
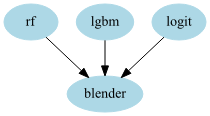
- Remark:
- the 2 models are equivalents
- to have regular stacking the same cvs should be used every where (either by creating it before hand, by setting the random state or using a non shuffle cv)
- With this idea we can do more complicated things like :
- deep stacking with more than one layer : simply add other layer to the GraphPipeline
- create a blender that uses both predictions of models as well as the features (or part of it) : simply add another node linked to the blender (a PassThrough node for example)
- do pre-processing before doing any stacking (and so doing it out-side of the cv loop)
For example:
stacker = GraphPipeline(models = {
"rf" : OutSamplerTransformer(RandomForestClassifier() , cv = cv),
"lgbm" : OutSamplerTransformer(LGBMClassifier() , cv = cv),
"logit": OutSamplerTransformer(LogisticRegression(), cv = cv),
"pass" : PassThrough(),
"blender":LogisticRegression()
}, edges = [("rf","blender"),
("lgbm","blender"),
("logit","blender"),
("pass", "blender")
])
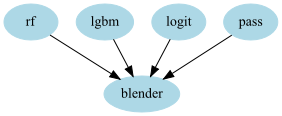
Or:
stacker = GraphPipeline(models = {
"enc" : NumericalEncoder(),
"imp" : NumImputer(),
"rf" : OutSamplerTransformer(RandomForestClassifier() , cv = cv),
"lgbm" : OutSamplerTransformer(LGBMClassifier() , cv = cv),
"logit": OutSamplerTransformer(LogisticRegression(), cv = cv),
"blender":LogisticRegression()
}, edges = [("enc","imp"),
("imp","rf","blender"),
("imp","lgbm","blender"),
("imp","logit","blender")
])
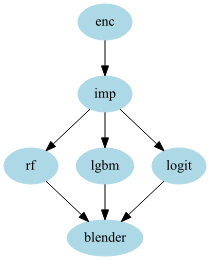
And lastly:
stacker = GraphPipeline(models = {
"enc" : NumericalEncoder(columns_to_use= ["cat1","cat2","num1","num2"]),
"imp" : NumImputer(),
"cv" : CountVectorizerWrapper(columns_to_use = ["text1","text2"]),
"logit": OutSamplerTransformer(LogisticRegression(), cv = cv),
"lgbm" : OutSamplerTransformer(LGBMClassifier() , cv = cv),
"blender":LogisticRegression()
}, edges = [("enc","imp","lgbm","blender"),
("cv","logit","blender")
])
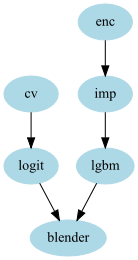
- This last example shows a model where you have:
- categorical and numerical data, on which you apply a classical categorie encoder, you fill missing value and use gradient boosting
- textual data which you can encode using a CountVectorizer and use LogisticRegression
Both models can be mixted with a Logistic Regression blending. Doing that just create an average between the predictions, the only difference is that since the blender is fitted, weights are in a sence optimal. (In some cases it might work better than just concatenate everything : especially since the 2 sets of features are highly different).
- Another thing that can be done is to calibrate probabilities. This can be useful if your model generate meaningful scores but you can’t directly interprete those scores as probabilities:
- if you skewed your training set to solve imbalance
- if your model is not probabilistic
- …
One method to re-calibrate probabilities, call Platt’s scaling , is to fit a LogisticRegression on the output of your model. If your predictions are not completely wrong, this will usualy just compute an increasing function that recalibrate your probabilities but won’t change change the ordering of the output. (roc_auc won’t be affected, but logloss, or accuracy can change).
This can also be done using OutSamplerTransformer:
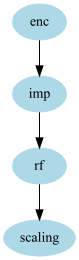
rf_rescaled = GraphPipeline(models = {
"enc" : NumericalEncoder(),
"imp" : NumImputer(),
"rf" : OutSamplerTransformer( RandomForestClassifier(class_weight = "auto"), cv = 10),
"scaling":LogisticRegression()
}, edges = [('enc','imp','rf','scaling')]
)
OutSamplerTransformer regression mode¶
You can do exactly the same thing but for regresion tasks, only difference is that cross-validation uses predict() instead of predict_proba().